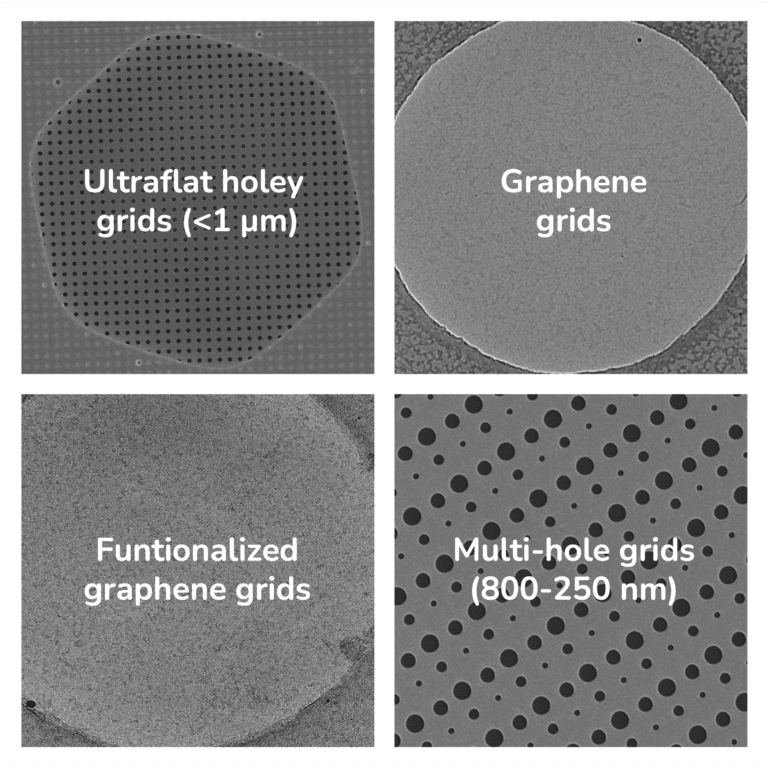
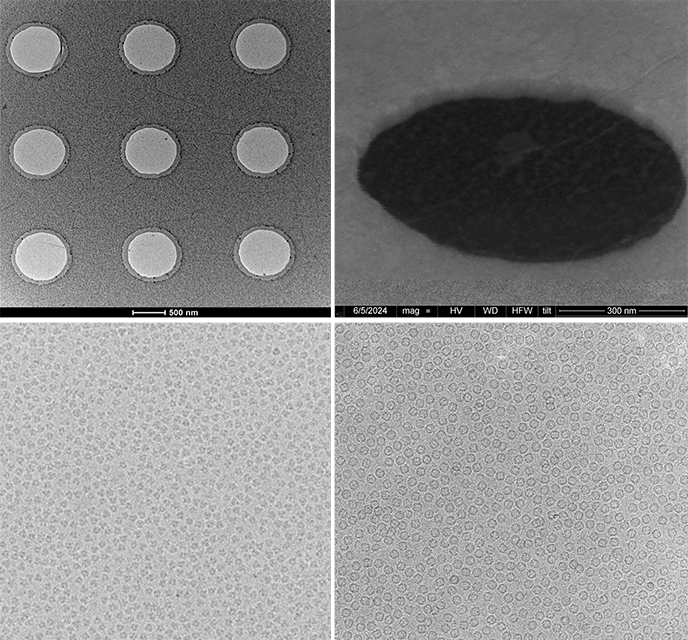
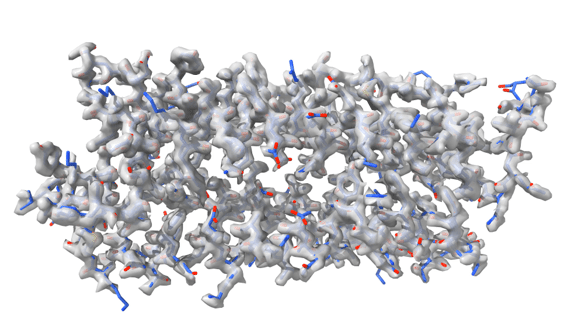
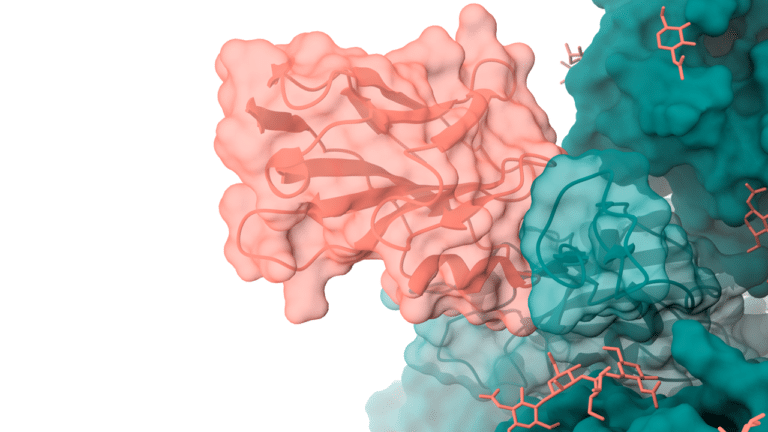
To provide the best experience, we use technologies such as cookies to store and/or access information about your device. By consenting to these technologies, we may process data such as browsing habits or unique IDs on this site. If you do not consent or withdraw your consent, certain features and capabilities may be adversely affected.
De technische opslag of toegang is strikt noodzakelijk voor het legitieme doel het gebruik mogelijk te maken van een specifieke dienst waarom de abonnee of gebruiker uitdrukkelijk heeft gevraagd, of met als enig doel de uitvoering van de transmissie van een communicatie over een elektronisch communicatienetwerk.
De technische opslag of toegang is noodzakelijk voor het legitieme doel voorkeuren op te slaan die niet door de abonnee of gebruiker zijn aangevraagd.
The technical storage or access is strictly necessary for the legitimate purpose of enabling the use of a specific service expressly requested by the subscriber or user, or for the sole purpose of carrying out the transmission of a communication over an electronic communications network.
De technische opslag of toegang die uitsluitend wordt gebruikt voor anonieme statistische doeleinden. Zonder dagvaarding, vrijwillige naleving door uw Internet Service Provider, of aanvullende gegevens van een derde partij, kan informatie die alleen voor dit doel wordt opgeslagen of opgehaald gewoonlijk niet worden gebruikt om u te identificeren.
The technical storage or access is necessary to create user profiles for sending advertising, or to track the user on a website or across different websites for similar marketing purposes.